Fine-tuning¶
FYI, you can open this documentation as a Google Colab notebook to follow along interactively
graph-pes provides access to several foundation models (with more being added as they are released) for you to use as-is, as well as to fine-tune on your own dataset. See the documentation for a growing list of the interfaces we provide.
Below, we:
install the relevant packages to use the MACE-MP-0 and Orb families of models
fine-tune the model on structures labelled with a different level of theory
Since graph-pes provides a unified interface to many different foundation models, swapping between these different foundation models is incredibly simple!
Installation¶
To make use of the MACE-MP-0 and Orb families of models, we need to install mace-torch and orb-models alongside graph-pes:
[ ]:
!pip install graph-pes mace-torch orb-models
Loading and using foundation models¶
Let’s start by loading the MACE-MP-0-small model and using it to make predictions on some SiO\(_2\) structures. To do this, we’ll use load-atoms to download the SiO2-GAP-22 dataset.
[1]:
from load_atoms import load_dataset
dataset = load_dataset("SiO2-GAP-22")
dataset
[1]:
SiO2-GAP-22:
structures: 3,074
atoms: 268,118
species:
O: 66.47%
Si: 33.53%
properties:
per atom: (forces)
per structure: (config_type, energy, stress, virial)
Let’s select the first structre from this dataset and visualise it:
[2]:
from load_atoms import view
structure = dataset[0]
view(structure, show_bonds=True)
[2]:
Now that we have a structure, lets use the graph_pes.interfaces.mace_mp function to load the MACE-MP-0-small model to make some predictions:
[3]:
import torch
from graph_pes.interfaces import mace_mp
device = "cuda" if torch.cuda.is_available() else "cpu"
print(f"Using device: {device}")
mp0 = mace_mp("small").eval().to(device)
sum(p.numel() for p in mp0.parameters())
Using device: cpu
[3]:
3847696
This model object is a GraphPESModel instance, and so can be used throughout the rest of the graph-pes ecosystem. For instance, we can inspect the dimer curves for this model:
[4]:
from graph_pes.utils.analysis import dimer_curve
import matplotlib.pyplot as plt
%config InlineBackend.figure_format = 'retina'
for dimer, c in [("Si-Si", "crimson"), ("O-O", "green"), ("Si-O", "royalblue")]:
dimer_curve(
mp0,
dimer.replace("-", ""),
units="eV",
rmin=0.7,
rmax=4.0,
label=dimer,
c=c,
)
plt.legend();
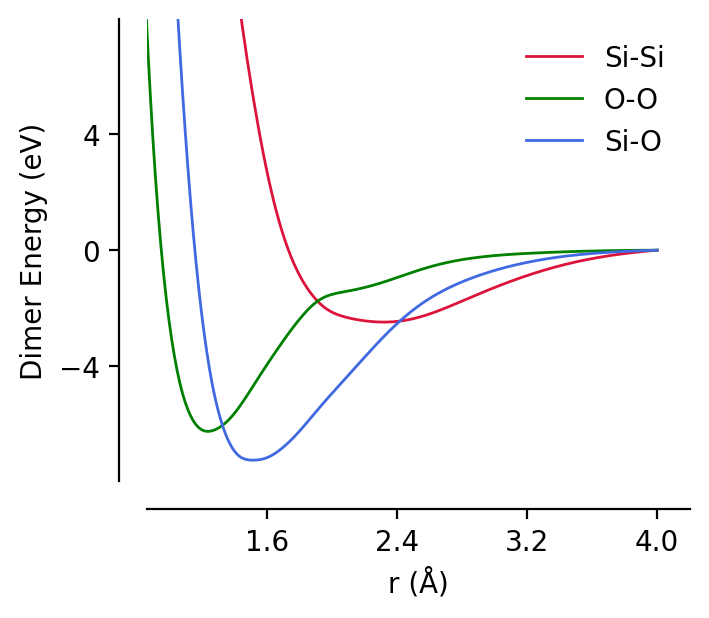
… and also use it as an ASE calculator to generate force predictions:
[5]:
from ase import Atoms
from graph_pes.graph_pes_model import GraphPESModel
def check_forces(model: GraphPESModel, structure: Atoms):
# make an ASE calculator
calculator = model.ase_calculator()
force_predictions = calculator.get_forces(structure)
force_labels = structure.arrays["forces"]
plt.figure(figsize=(2.5, 2.5))
plt.scatter(force_labels.flatten(), force_predictions.flatten(), s=6)
plt.axline((0, 0), slope=1, color="black", ls="--", lw=1)
plt.gca().set_aspect("equal")
plt.xlabel("True force / eV/Å")
plt.ylabel("Predicted force / eV/Å")
check_forces(mp0, dataset[0])
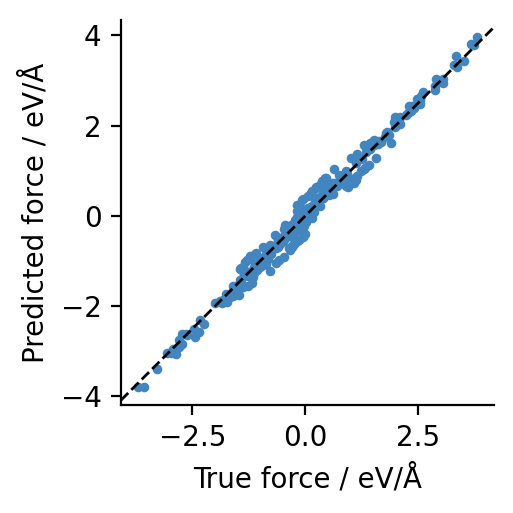
To demonstrate the architecture-agnostic nature of graph-pes, we can swap between the MACE-MP-0-small and Orb-v3-small models very easily:
[6]:
from graph_pes.interfaces import orb_model
orbv2_xs = orb_model("orb-d3-xs-v2").eval().to(device)
sum(p.numel() for p in orbv2_xs.parameters())
[6]:
9443981
[7]:
check_forces(orbv2_xs, dataset[0])
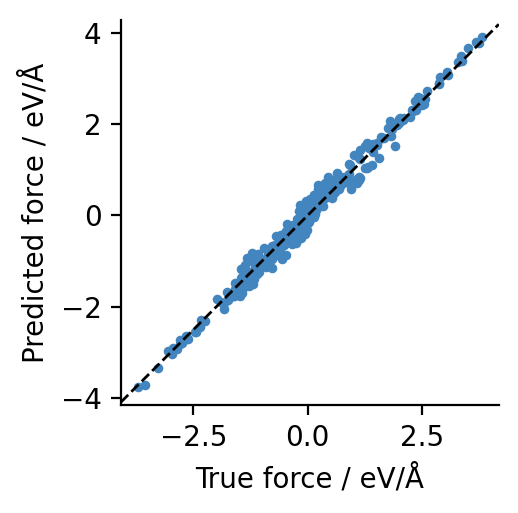
It is explicitly not the point here to compare these two models. Instead, we are just demonstrating that these two models, created by different teams and originating from different packages, can be treated identically within the confines of graph-pes.
We now know how to use these foundation models out of the box - they behave just like any other graph-pes model!
Fine-tuning¶
Often we want to fine-tune foundation models on specific datasets to further improve their accuracy. graph-pes makes this easy!
We saw above that the force predictions for the foundation models were already in close agreement with the DFT labels of the SiO2-GAP-22 model. This is despite the fact that these foundation models were trained on data labelled with different functionals.
However, different functionals tend to have different “reference” energies, i.e., the energy of an isolated atom of element \(X\) will have some non-0 energy, \(\varepsilon_X\) and for functional (a) vs (b), \(\varepsilon_X^{(a)} \neq \varepsilon_X^{(b)}\).
To demonstrate this, we create an energy parity plot below:
[8]:
from graph_pes.utils.analysis import parity_plot
parity_plot(
mp0,
dataset[:20],
property="energy_per_atom",
units="eV/atom",
c="crimson",
label="MP0",
)
parity_plot(
orbv2_xs,
dataset[:20],
property="energy_per_atom",
units="eV/atom",
c="royalblue",
label="Orb",
)
plt.xlabel("Ground truth SCAN Energy (eV/atom)")
plt.ylabel("Predicted Energy (eV/atom)")
plt.legend(frameon=True);
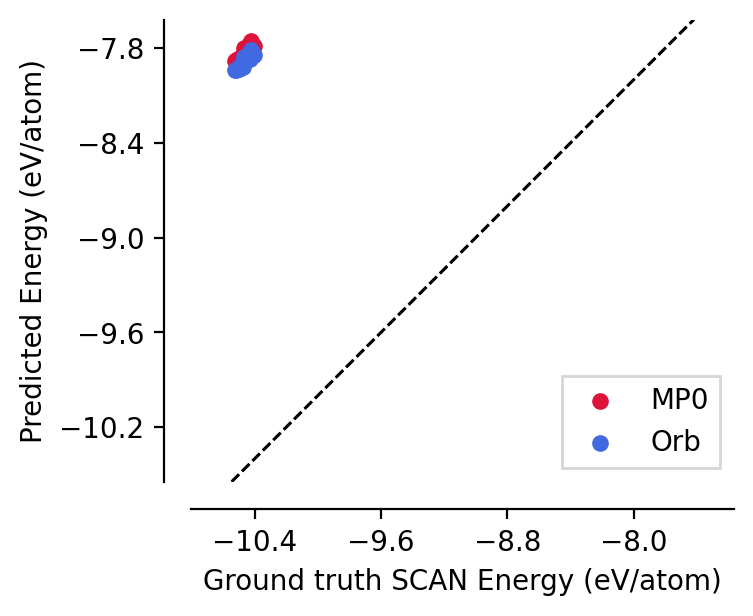
graph-pes provides automated functionality to correct for exactly these kind of differences:
[9]:
from graph_pes.utils.shift_and_scale import add_auto_offset
adjusted_mp0 = add_auto_offset(mp0, dataset[:20])
adjusted_orb = add_auto_offset(orbv2_xs, dataset[:20])
parity_plot(
adjusted_mp0,
dataset[:20],
property="energy_per_atom",
units="eV/atom",
c="crimson",
label="MP0",
)
parity_plot(
adjusted_orb,
dataset[:20],
property="energy_per_atom",
units="eV/atom",
c="royalblue",
label="Orb",
)
plt.xlabel("Ground truth SCAN Energy (eV/atom)")
plt.ylabel("Predicted Energy (eV/atom)")
plt.legend(frameon=True);
[graph-pes INFO]:
Attempting to automatically detect the offset energy for each element.
We do this by first generating predictions for each training structure
(up to `config.fitting.max_n_pre_fit` if specified).
This is a slow process! If you already know the reference energies (or the
difference in reference energies if you are fine-tuning an existing model to a
different level of theory),
we recommend setting `config.fitting.auto_fit_reference_energies` to `False`
and manually specifying a `LearnableOffset` component of your model.
See the "Fine-tuning" tutorial in the docs for more information:
https://jla-gardner.github.io/graph-pes/quickstart/fine-tuning.html
[graph-pes WARNING]:
We are attempting to guess the mean per-element
contribution for a per-structure quantity (usually
the total energy).
However, the composition of the training set is such that
no unique solution is possible.
This is probably because you are training on structures
all with the same composition (e.g. all structures are
of the form n H2O). Consider explicitly setting the
per-element contributions if you know them, or
including a variety of structures of different
compositions in the training set.
[graph-pes INFO]:
Attempting to automatically detect the offset energy for each element.
We do this by first generating predictions for each training structure
(up to `config.fitting.max_n_pre_fit` if specified).
This is a slow process! If you already know the reference energies (or the
difference in reference energies if you are fine-tuning an existing model to a
different level of theory),
we recommend setting `config.fitting.auto_fit_reference_energies` to `False`
and manually specifying a `LearnableOffset` component of your model.
See the "Fine-tuning" tutorial in the docs for more information:
https://jla-gardner.github.io/graph-pes/quickstart/fine-tuning.html
[graph-pes WARNING]:
We are attempting to guess the mean per-element
contribution for a per-structure quantity (usually
the total energy).
However, the composition of the training set is such that
no unique solution is possible.
This is probably because you are training on structures
all with the same composition (e.g. all structures are
of the form n H2O). Consider explicitly setting the
per-element contributions if you know them, or
including a variety of structures of different
compositions in the training set.
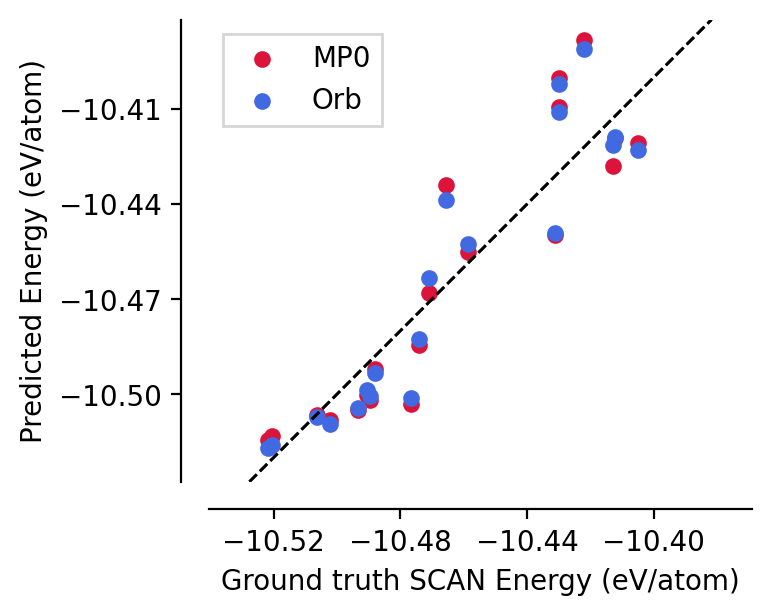
Note the warning above! It we pass a set of structures with exactly the same composition (here \(n\cdot SiO_2\)), its not possible to decouple the differences in the \(\varepsilon_O\) and \(\varepsilon_{Si}\). This means if you use this new model to predict energies on a new structure with a different compositions, e.g. \(Si_5O_9\), you will get very different energies from what the ground truth can give you.
With that in mind, lets get down to fine-tuning a model. As in our other fine-tuning guide we start by selecting some structures to fine-tune on:
[10]:
train, valid = dataset[:20], dataset[20:25]
train.write("train.xyz")
valid.write("valid.xyz")
Next, we define a config file to describe how what and how we want to fine-tune. Two vital features here are:
the
fitting/auto_fit_reference_energies=trueflag - this performs the above automated offset guessing before any fine-tuning takes placeusing a relatively small learning rate
[20]:
accelerator = "gpu" if torch.cuda.is_available() else "cpu"
training_config = f"""
data:
train: train.xyz
valid: valid.xyz
loss:
- +PerAtomEnergyLoss()
- +ForceRMSE()
fitting:
trainer_kwargs:
max_epochs: 20
accelerator: {accelerator}
optimizer:
name: Adam
lr: 0.0001
auto_fit_reference_energies: true
wandb: null
general:
progress: logged
run_id: mp0-fine-tune
"""
mp0_config = """
model:
+mace_mp:
model: small
general:
run_id: mp0-fine-tune
"""
with open("fine-tune.yaml", "w") as f:
f.write(training_config)
with open("mp0.yaml", "w") as f:
f.write(mp0_config)
Now we can fine-tune. Note the use of config file stacking here - this lets us separate the config into different files, which can often be useful! (see below)
[12]:
!graph-pes-train fine-tune.yaml mp0.yaml
[graph-pes INFO]: Started `graph-pes-train` at 2025-04-11 16:52:15.879
[graph-pes INFO]: Successfully parsed config.
[graph-pes INFO]: Logging to graph-pes-results/mp0-fine-tune/rank-0.log
[graph-pes INFO]: ID for this training run: mp0-fine-tune
[graph-pes INFO]:
Output for this training run can be found at:
└─ graph-pes-results/mp0-fine-tune
├─ rank-0.log # find a verbose log here
├─ model.pt # the best model (according to valid/loss/total)
├─ lammps_model.pt # the best model deployed to LAMMPS
├─ train-config.yaml # the complete config used for this run
└─ summary.yaml # the summary of the training run
GPU available: True (mps), used: False
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
[graph-pes INFO]: Preparing data
[graph-pes INFO]: Setting up datasets
[graph-pes INFO]: Pre-fitting the model on 20 samples
[graph-pes INFO]:
Attempting to automatically detect the offset energy for each element.
We do this by first generating predictions for each training structure
(up to `config.fitting.max_n_pre_fit` if specified).
This is a slow process! If you already know the reference energies (or the
difference in reference energies if you are fine-tuning an existing model to a
different level of theory),
we recommend setting `config.fitting.auto_fit_reference_energies` to `False`
and manually specifying a `LearnableOffset` component of your model.
See the "Fine-tuning" tutorial in the docs for more information:
https://jla-gardner.github.io/graph-pes/quickstart/fine-tuning.html
[graph-pes WARNING]:
We are attempting to guess the mean per-element
contribution for a per-structure quantity (usually
the total energy).
However, the composition of the training set is such that
no unique solution is possible.
This is probably because you are training on structures
all with the same composition (e.g. all structures are
of the form n H2O). Consider explicitly setting the
per-element contributions if you know them, or
including a variety of structures of different
compositions in the training set.
[graph-pes INFO]:
Number of learnable params:
base (MACEWrapper) : 3,847,696
auto_offset (LearnableOffset): 2
[graph-pes INFO]: Sanity checking the model...
[graph-pes INFO]: Starting fit...
valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics timer/its_per_s timer/its_per_s
epoch time per_atom_energy_rmse per_atom_energy_mae energy_rmse energy_mae forces_rmse forces_mae stress_rmse stress_mae virial_rmse virial_mae train valid
1 7.8 0.00716 0.00673 0.77302 0.72710 0.10150 0.07997 0.00925 0.00584 128.09291 102.27287 0.80064 4.89904
2 16.1 0.00905 0.00825 0.97704 0.89060 0.05779 0.04414 0.00524 0.00346 128.24873 103.04964 0.70522 4.23109
3 24.6 0.00174 0.00132 0.18810 0.14233 0.03947 0.02871 0.00277 0.00183 128.95222 103.92886 0.64683 4.50949
4 32.7 0.00432 0.00419 0.46684 0.45242 0.03426 0.02702 0.00179 0.00117 130.01169 104.82194 0.71276 4.59975
5 40.1 0.00162 0.00142 0.17489 0.15320 0.03540 0.02763 0.00139 0.00091 130.51797 105.21088 0.78493 4.31182
6 48.0 0.00332 0.00312 0.35861 0.33645 0.02760 0.02140 0.00186 0.00125 129.84111 104.61426 0.67797 4.33375
7 56.0 0.00225 0.00195 0.24346 0.21077 0.02652 0.02051 0.00180 0.00125 129.79610 104.55513 0.67476 4.14495
8 64.0 0.00140 0.00090 0.15127 0.09751 0.02467 0.01905 0.00183 0.00127 129.90742 104.63938 0.75873 3.79053
9 71.9 0.00206 0.00177 0.22223 0.19067 0.02435 0.01856 0.00168 0.00113 130.40732 105.05786 0.75075 3.73735
10 79.7 0.00240 0.00231 0.25874 0.24932 0.02326 0.01803 0.00110 0.00075 130.71355 105.33549 0.68776 4.25472
11 87.3 0.00208 0.00199 0.22486 0.21538 0.02311 0.01779 0.00106 0.00070 130.88290 105.47400 0.76104 4.32935
12 95.0 0.00122 0.00101 0.13136 0.10957 0.02261 0.01729 0.00115 0.00075 130.93184 105.50145 0.78802 4.60993
13 101.1 0.00068 0.00040 0.07350 0.04363 0.02162 0.01665 0.00108 0.00072 130.93443 105.49496 1.02987 5.04530
14 107.4 0.00062 0.00047 0.06713 0.05095 0.02094 0.01612 0.00106 0.00070 131.02118 105.55566 0.76511 6.38889
15 114.5 0.00092 0.00078 0.09890 0.08396 0.02081 0.01601 0.00087 0.00058 131.20523 105.70125 0.77821 6.11329
16 123.3 0.00220 0.00217 0.23743 0.23433 0.02170 0.01662 0.00069 0.00045 131.44841 105.88866 0.65963 3.55058
17 132.7 0.00166 0.00158 0.17911 0.17083 0.01936 0.01502 0.00088 0.00059 131.26625 105.73106 0.54230 3.37123
18 140.1 0.00130 0.00126 0.14066 0.13652 0.01936 0.01496 0.00062 0.00042 131.49023 105.90979 0.76336 4.44092
19 147.6 0.00111 0.00109 0.12030 0.11782 0.02065 0.01572 0.00047 0.00031 131.74437 106.11167 0.75529 4.32136
20 155.2 0.00304 0.00303 0.32831 0.32720 0.01846 0.01435 0.00045 0.00033 131.59532 105.98393 0.80128 5.05800
`Trainer.fit` stopped: `max_epochs=20` reached.
[graph-pes INFO]: Loading best weights from "/Users/john/projects/graph-pes/docs/source/quickstart/graph-pes-results/mp0-fine-tune/checkpoints/best.ckpt"
[graph-pes INFO]: Training complete.
[graph-pes INFO]: Testing best model...
GPU available: True (mps), used: False
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Test metric ┃ DataLoader 0 ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ test/train/energy_mae │ 0.08073119819164276 │
│ test/train/energy_rmse │ 0.09959671646356583 │
│ test/train/forces_mae │ 0.014654574915766716 │
│ test/train/forces_rmse │ 0.0190340057015419 │
│ test/train/forces_rmse_batchwise │ 0.018963992595672607 │
│ test/train/per_atom_energy_mae │ 0.0007476329919882119 │
│ test/train/per_atom_energy_rmse │ 0.0009222375811077654 │
│ test/train/stress_mae │ 0.00044360198080539703 │
│ test/train/stress_rmse │ 0.0006201165379025042 │
│ test/train/virial_mae │ 97.18195343017578 │
│ test/train/virial_rmse │ 127.16856384277344 │
│ test/valid/energy_mae │ │
│ test/valid/energy_rmse │ │
│ test/valid/forces_mae │ │
│ test/valid/forces_rmse │ │
│ test/valid/forces_rmse_batchwise │ │
│ test/valid/per_atom_energy_mae │ │
│ test/valid/per_atom_energy_rmse │ │
│ test/valid/stress_mae │ │
│ test/valid/stress_rmse │ │
│ test/valid/virial_mae │ │
│ test/valid/virial_rmse │ │
└──────────────────────────────────┴──────────────────────────────────┘
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Test metric ┃ DataLoader 1 ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ test/train/energy_mae │ │
│ test/train/energy_rmse │ │
│ test/train/forces_mae │ │
│ test/train/forces_rmse │ │
│ test/train/forces_rmse_batchwise │ │
│ test/train/per_atom_energy_mae │ │
│ test/train/per_atom_energy_rmse │ │
│ test/train/stress_mae │ │
│ test/train/stress_rmse │ │
│ test/train/virial_mae │ │
│ test/train/virial_rmse │ │
│ test/valid/energy_mae │ 0.13652344048023224 │
│ test/valid/energy_rmse │ 0.10905405879020691 │
│ test/valid/forces_mae │ 0.014955026097595692 │
│ test/valid/forces_rmse │ 0.01909947581589222 │
│ test/valid/forces_rmse_batchwise │ 0.019314901903271675 │
│ test/valid/per_atom_energy_mae │ 0.001264381455257535 │
│ test/valid/per_atom_energy_rmse │ 0.0010098782368004322 │
│ test/valid/stress_mae │ 0.00042308951378799975 │
│ test/valid/stress_rmse │ 0.0006206676480360329 │
│ test/valid/virial_mae │ 105.9097900390625 │
│ test/valid/virial_rmse │ 128.0445556640625 │
└──────────────────────────────────┴──────────────────────────────────┘
[graph-pes INFO]: Testing complete.
[graph-pes INFO]: Awaiting final Lightning and W&B shutdown...
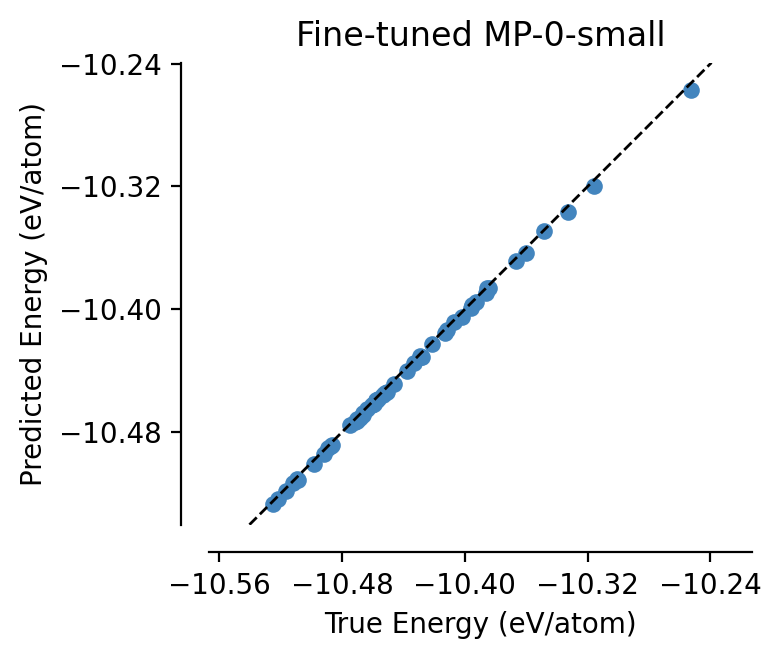
Nice! Fine-tuning on just 20 structures has brought the validation error down significantly 😊 Of course in real life, one would train for longer, and perhaps make use of early stopping to prevent overfitting to the small amounts of data - see the docs for the relevant config options you need to pass to do that.
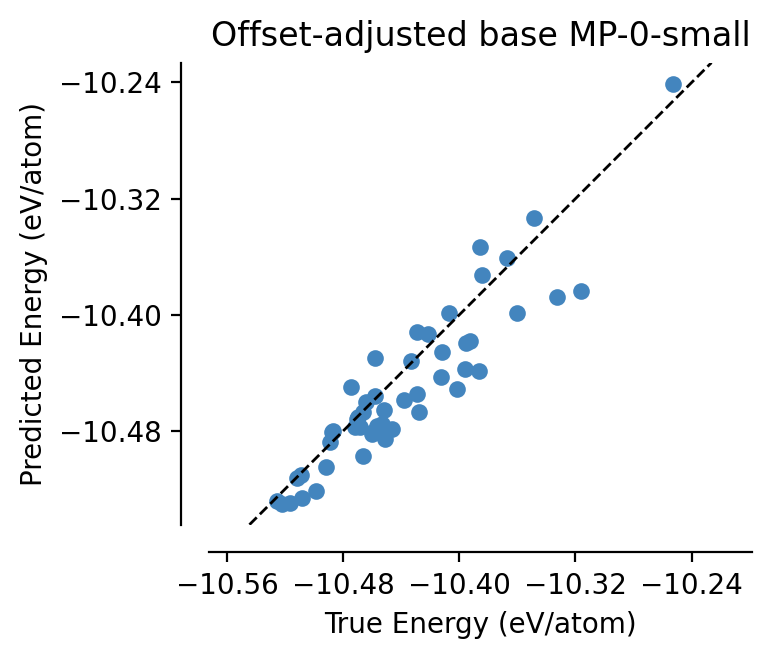
Lets load in our fine-tuned model and check that everything is working as expected:
[13]:
from graph_pes.models import load_model
test_set = dataset[50:100]
parity_plot(
adjusted_mp0,
test_set,
property="energy_per_atom",
units="eV/atom",
)
plt.title("Offset-adjusted base MP-0-small")
plt.show()
fine_tuned_mp0 = load_model("graph-pes-results/mp0-fine-tune/model.pt").eval()
parity_plot(
fine_tuned_mp0,
test_set,
property="energy_per_atom",
units="eV/atom",
)
plt.title("Fine-tuned MP-0-small")
[13]:
Text(0.5, 1.0, 'Fine-tuned MP-0-small')
Swapping out the MP-0 model for the Orb model is trivial! We just need to change the model definition in the config file. Here we’re being extra fancy, and freezing all the model parameters except those in the read-out head (see graph_pes.models.freeze_all_except for details):
[24]:
orb_config = """
model:
+freeze_all_except:
model:
+orb_model:
name: orb-d3-xs-v2
pattern: _orb\\.heads.*
general:
run_id: orb-fine-tune
"""
with open("orb.yaml", "w") as f:
f.write(orb_config)
[25]:
!graph-pes-train fine-tune.yaml orb.yaml
/opt/miniconda3/envs/mace/lib/python3.11/site-packages/e3nn/o3/_wigner.py:10: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
_Jd, _W3j_flat, _W3j_indices = torch.load(os.path.join(os.path.dirname(__file__), 'constants.pt'))
[graph-pes INFO]: Started `graph-pes-train` at 2025-04-11 17:16:02.923
/opt/miniconda3/envs/mace/lib/python3.11/site-packages/orb_models/utils.py:30: UserWarning: Setting global torch default dtype to torch.float32.
warnings.warn(f"Setting global torch default dtype to {torch_dtype}.")
[graph-pes INFO]: Successfully parsed config.
[graph-pes INFO]: Logging to graph-pes-results/orb-fine-tune/rank-0.log
[graph-pes INFO]: ID for this training run: orb-fine-tune
[graph-pes INFO]:
Output for this training run can be found at:
└─ graph-pes-results/orb-fine-tune
├─ rank-0.log # find a verbose log here
├─ model.pt # the best model (according to valid/loss/total)
├─ lammps_model.pt # the best model deployed to LAMMPS
├─ train-config.yaml # the complete config used for this run
└─ summary.yaml # the summary of the training run
GPU available: True (mps), used: False
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
[graph-pes INFO]: Preparing data
[graph-pes INFO]: Setting up datasets
[graph-pes INFO]: Pre-fitting the model on 20 samples
[graph-pes INFO]:
Attempting to automatically detect the offset energy for each element.
We do this by first generating predictions for each training structure
(up to `config.fitting.max_n_pre_fit` if specified).
This is a slow process! If you already know the reference energies (or the
difference in reference energies if you are fine-tuning an existing model to a
different level of theory),
we recommend setting `config.fitting.auto_fit_reference_energies` to `False`
and manually specifying a `LearnableOffset` component of your model.
See the "Fine-tuning" tutorial in the docs for more information:
https://jla-gardner.github.io/graph-pes/quickstart/fine-tuning.html
[graph-pes WARNING]:
We are attempting to guess the mean per-element
contribution for a per-structure quantity (usually
the total energy).
However, the composition of the training set is such that
no unique solution is possible.
This is probably because you are training on structures
all with the same composition (e.g. all structures are
of the form n H2O). Consider explicitly setting the
per-element contributions if you know them, or
including a variety of structures of different
compositions in the training set.
[graph-pes INFO]:
Number of learnable params:
base (OrbWrapper) : 200,064
auto_offset (LearnableOffset): 2
[graph-pes INFO]: Sanity checking the model...
[graph-pes INFO]: Starting fit...
valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics valid/metrics timer/its_per_s timer/its_per_s
epoch time per_atom_energy_rmse per_atom_energy_mae energy_rmse energy_mae forces_rmse forces_mae stress_rmse stress_mae virial_rmse virial_mae train valid
1 1.8 0.01108 0.00818 1.19668 0.88313 0.16017 0.12256 0.02922 0.02326 76.28781 61.23480 3.80228 5.45729
2 3.6 0.00770 0.00585 0.83182 0.63171 0.15506 0.11872 0.02922 0.02326 76.28781 61.23480 4.13223 11.88725
3 5.3 0.00467 0.00438 0.50436 0.47307 0.15038 0.11539 0.02922 0.02326 76.28781 61.23480 3.86100 10.40802
4 7.1 0.00369 0.00305 0.39796 0.32947 0.14623 0.11259 0.02922 0.02326 76.28781 61.23480 4.03226 10.95286
5 8.9 0.00262 0.00232 0.28348 0.25066 0.14247 0.11010 0.02922 0.02326 76.28781 61.23480 3.04878 10.65872
6 10.7 0.00185 0.00136 0.19947 0.14702 0.13893 0.10778 0.02922 0.02326 76.28781 61.23480 4.42478 11.21332
7 12.5 0.00217 0.00192 0.23384 0.20730 0.13528 0.10521 0.02922 0.02326 76.28781 61.23480 3.90625 11.81803
8 14.1 0.00266 0.00254 0.28749 0.27485 0.13179 0.10278 0.02922 0.02326 76.28781 61.23480 4.31034 10.62050
9 16.0 0.00250 0.00214 0.27035 0.23108 0.12831 0.10037 0.02922 0.02326 76.28781 61.23480 3.80228 8.25962
10 17.8 0.00269 0.00243 0.29018 0.26196 0.12483 0.09784 0.02922 0.02326 76.28781 61.23480 4.52489 10.93097
11 19.5 0.00218 0.00198 0.23518 0.21355 0.12133 0.09523 0.02922 0.02326 76.28781 61.23480 4.40529 11.30330
12 21.1 0.00259 0.00245 0.27953 0.26460 0.11804 0.09287 0.02922 0.02326 76.28781 61.23480 4.36681 11.35335
13 22.8 0.00248 0.00226 0.26789 0.24412 0.11474 0.09049 0.02922 0.02326 76.28781 61.23480 4.08163 11.98733
14 24.4 0.00215 0.00185 0.23196 0.19985 0.11158 0.08824 0.02922 0.02326 76.28781 61.23480 3.42466 11.44267
15 26.0 0.00353 0.00302 0.38135 0.32583 0.10840 0.08593 0.02922 0.02326 76.28781 61.23480 3.41297 11.50141
16 27.7 0.00368 0.00289 0.39733 0.31230 0.10509 0.08339 0.02922 0.02326 76.28781 61.23480 3.48432 11.62694
17 29.4 0.00328 0.00277 0.35471 0.29954 0.10226 0.08139 0.02922 0.02326 76.28781 61.23480 4.42478 9.53071
18 31.1 0.00258 0.00209 0.27859 0.22561 0.09923 0.07914 0.02922 0.02326 76.28781 61.23480 2.79330 11.77056
19 32.8 0.00365 0.00313 0.39438 0.33806 0.09616 0.07678 0.02922 0.02326 76.28781 61.23480 2.94118 10.99205
20 34.5 0.00269 0.00218 0.29059 0.23569 0.09309 0.07437 0.02922 0.02326 76.28781 61.23480 4.00000 10.98432
`Trainer.fit` stopped: `max_epochs=20` reached.
[graph-pes INFO]: Loading best weights from "/Users/john/projects/graph-pes/docs/source/quickstart/graph-pes-results/orb-fine-tune/checkpoints/best.ckpt"
[graph-pes INFO]: Training complete.
[graph-pes INFO]: Testing best model...
GPU available: True (mps), used: False
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Test metric ┃ DataLoader 0 ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ test/train/energy_mae │ 0.16972656548023224 │
│ test/train/energy_rmse │ 0.23350417613983154 │
│ test/train/forces_mae │ 0.07444514334201813 │
│ test/train/forces_rmse │ 0.09575104713439941 │
│ test/train/forces_rmse_batchwise │ 0.09573473781347275 │
│ test/train/per_atom_energy_mae │ 0.0015714168548583984 │
│ test/train/per_atom_energy_rmse │ 0.002161899348720908 │
│ test/train/stress_mae │ 0.02080981805920601 │
│ test/train/stress_rmse │ 0.0265880785882473 │
│ test/train/virial_mae │ 61.88399124145508 │
│ test/train/virial_rmse │ 92.37788391113281 │
│ test/valid/energy_mae │ │
│ test/valid/energy_rmse │ │
│ test/valid/forces_mae │ │
│ test/valid/forces_rmse │ │
│ test/valid/forces_rmse_batchwise │ │
│ test/valid/per_atom_energy_mae │ │
│ test/valid/per_atom_energy_rmse │ │
│ test/valid/stress_mae │ │
│ test/valid/stress_rmse │ │
│ test/valid/virial_mae │ │
│ test/valid/virial_rmse │ │
└──────────────────────────────────┴──────────────────────────────────┘
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Test metric ┃ DataLoader 1 ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ test/train/energy_mae │ │
│ test/train/energy_rmse │ │
│ test/train/forces_mae │ │
│ test/train/forces_rmse │ │
│ test/train/forces_rmse_batchwise │ │
│ test/train/per_atom_energy_mae │ │
│ test/train/per_atom_energy_rmse │ │
│ test/train/stress_mae │ │
│ test/train/stress_rmse │ │
│ test/train/virial_mae │ │
│ test/train/virial_rmse │ │
│ test/valid/energy_mae │ 0.23569336533546448 │
│ test/valid/energy_rmse │ 0.2459844946861267 │
│ test/valid/forces_mae │ 0.07436826825141907 │
│ test/valid/forces_rmse │ 0.09522523730993271 │
│ test/valid/forces_rmse_batchwise │ 0.09307248890399933 │
│ test/valid/per_atom_energy_mae │ 0.002182579133659601 │
│ test/valid/per_atom_energy_rmse │ 0.002277533058077097 │
│ test/valid/stress_mae │ 0.023262472823262215 │
│ test/valid/stress_rmse │ 0.027134379372000694 │
│ test/valid/virial_mae │ 61.23480224609375 │
│ test/valid/virial_rmse │ 89.39186096191406 │
└──────────────────────────────────┴──────────────────────────────────┘
[graph-pes INFO]: Testing complete.
[graph-pes INFO]: Awaiting final Lightning and W&B shutdown...
[26]:
fine_tuned_orb = load_model("graph-pes-results/orb-fine-tune/model.pt").eval()
parity_plot(
fine_tuned_orb,
test_set,
property="energy_per_atom",
units="eV/atom",
)
plt.title("Fine-tuned orb-d3-xs-v2");
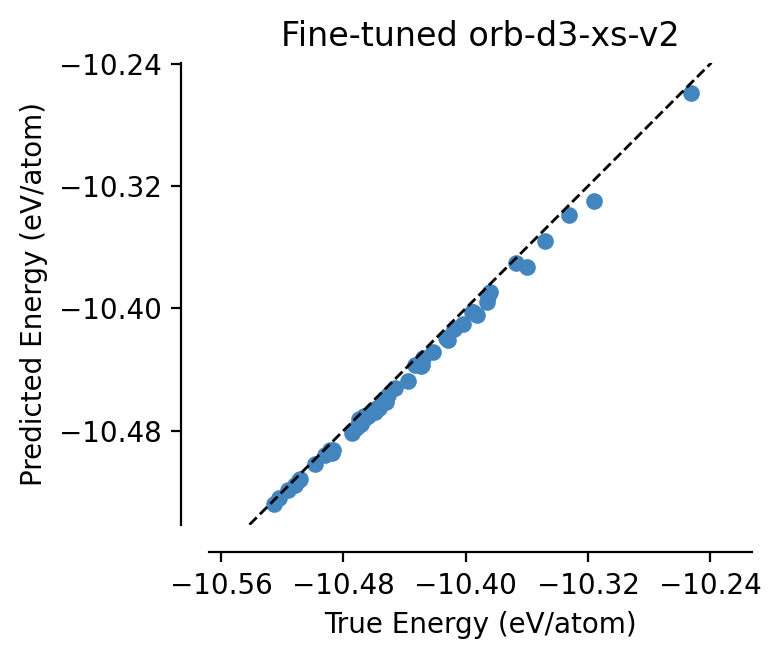
Again, we aren’t trying to make direct comparisons between these two models here - the instances we have chosen are both small models to show proof of concept.
If you want to explore fine-tuning various foundation models, we strongly recommend that you tune the hyperparameters of the fine-tuning process for each one separately.
Fine-tuning other models¶
Of course, you can also fine-tune other models that you have trained yourself! In this case, rather than using the foundation model interfaces, use the load_model function to load your model from disk. Remember to pass the auto_fit_reference_energies=true flag to the graph-pes-train command to ensure that this model correctly updates its reference energies:
model:
+load_model:
path: path/to/model.pt
fitting:
auto_fit_reference_energies: true
# ... <-- other config options as normal